Editorial by Prof Manolis Tsiknakis, Coordinator of ProCAncer-I project
ProCAncer-I project after three years of implementation
ProCAncer-I’s vision is to deliver a platform featuring a unique collection of PCa mpMRI images worldwide, in terms of data quantity, quality and diversity; to focus on delivering novel AI-based clinical tools for advancing characterization of PCa lesions, assessment of the metastatic potential, and early detection of disease recurrence; to design and seamlessly integrate an open source framework for the development, sharing and deployment of AI models and tools; to contribute in the increase of the trust in PCa AI tools by introducing AI model interpretability functionality and delivering a complete technological, organizational, and legislative framework for model evaluation; and to develop a concrete plan to sustain and exploit project result.
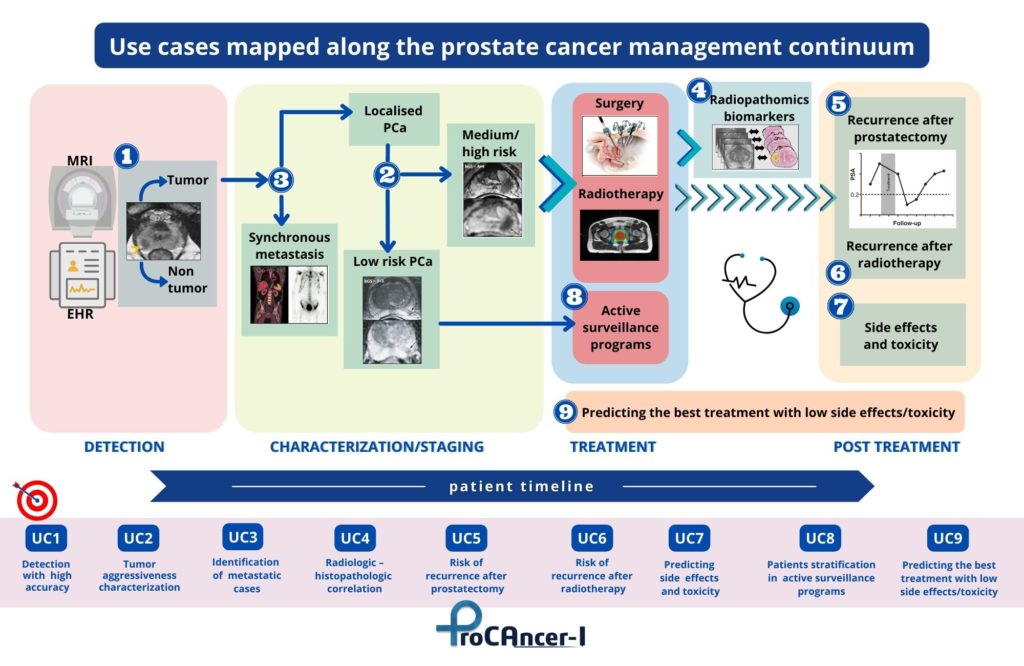
ProCAncer-I has developed nine concrete and clinically relevant use cases that span the care continuum of PCa. These are used as demonstrator use cases for validating the added value and usability of the platform. The ProCAncer-I project has achieved significant milestones from the clinical viewpoint, including the definition of detailed study protocols for retrospective and prospective studies, and is registered on clinicaltrial.gov. The project also established robust procedures and technologies for data anonymization, developed the ProCAncer-I platform as a secure cloud-based infrastructure supporting AI model development, and delivered locally installed eCRF and data upload tools to clinical partners, enhancing data control capabilities.
A significant focus of our work has been on dealing ontologies and catalog mechanisms. The MOLGENIS platform functions as the primary metadata catalogue, aligned with the DCAT-AP specification. Moreover, the project uses the OMOP-CDM, along with its extensions introduced within ProCAncer-I, as a common data model for storing clinical and imaging-related metadata. Collaboration with the OHDSI Medical Imaging Working Group persisted, focusing on integrating annotation, segmentation, and curation data as radiomics features, leading to the creation of two extensions to the OMOP-CDM (MI-CDM and R-CDM).
Substantial efforts were dedicated to enhancing the platform with various image pre-processing and curation tools including Bias Field Correction, Image Enhancement (RACLAHE histogram equalization, Deep Learning Noise Reduction, Radiomics Normalization (based on ComBat method) and Radiomic Feature Stability. Also, a significant result of our work has been the development of master models, as foundational models used for different tasks and methodologies. Efforts were directed mainly towards creating classification master models based on radiomics and deep learning (DL), alongside segmentation master models for whole prostate gland, prostate zone, and lesion segmentation. Certain partners concentrated on a consistent feature extraction and machine-learning pipeline using automatic whole prostate gland segmentations to evaluate predictive performance across various use cases (UC2, UC3, UC5, UC6, UC7b, and UC8). They conducted analyses like fairness, learning curve, and feature importance to understand diverse model requirements and feature impacts on performance. Other partners focused on analyzing performance using manually annotated lesion or whole prostate gland segmentation masks, offering a comparison between different model requirements, whereas others assessed radiomics features on UC7a, employing predicted whole prostate gland segmentations.
Regarding the development of Deep-Learning based Master Model, we have:
• Studied the impact of different factors on classification performance in UC2, including model types, clinical features, crop sizes, and data amounts.
• Studied the impact of image cropping techniques in UC2 and UC5 classification, alongside learning curve analysis.
• Compared unsupervised and supervised approaches in UC1, providing a learning curve analysis.
• Compared 2D and 3D data performances in prostate and lesion segmentation models.
• Examined architecture impacts on lesion segmentation models using mpMRI data.
• Enhanced existing prostate segmentation model’s robustness.
• Designed a deep learning-based lesion segmentation model and strategies to address overfitting and
• Investigated self-supervised learning (SSL) models’ performance in 3D classification using 2D orphan data stored in DICOM format, comparing their performance with models trained in previous chapters.
Ongoing experiments with deep learning models highlight benefits and dependencies on data quantity, case complexity, and image quality variations, offering adaptable AI strategies for better models addressing clinical questions. The project is collaborating with AI4HI network projects and notably, it contributed to defining “FUTURE-AI” principles, essential for AI model development in medical imaging and generated significant scientific output, publishing AI-based model reports.

