by Quibim
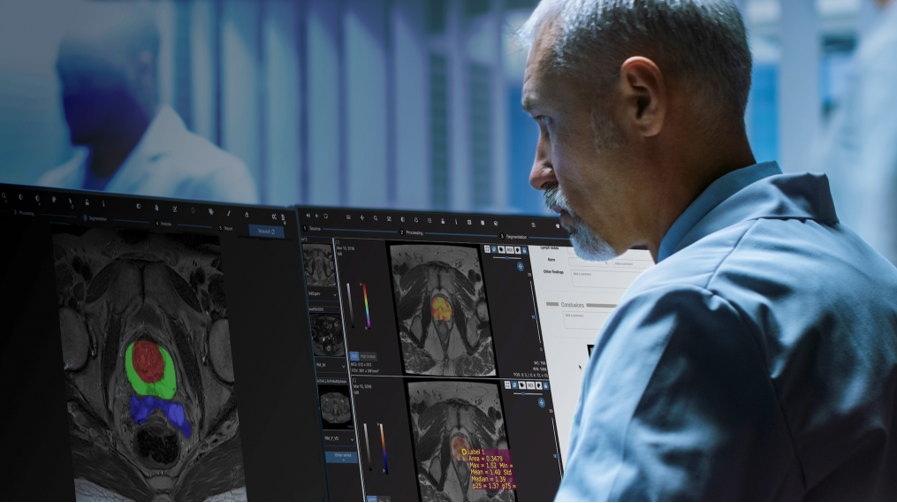
Quibim set out to transform prostate diagnosis and monitoring by developing a new non-invasive imaging tool using MRI data and advanced computer models to investigate the prostate anatomy in extreme detail. The company has developed AI algorithms for segmentation of prostate gland (peripheral zone, central gland, seminal vesicles) with PI-RADS 2.1 parcellation that can be modified by the experts to store verified annotations and that will be made available to the project. The annotation tool allows to have a concurrent annotation from multiple users, keeping an audit trail that makes secure and reliable use of this data.
Quibim is in parallel implementing 3 tools and models at the ProCAncer-I project are the following:
• Implementing an image and data annotation tool.
• Developing a system for the monitoring, loging, and retraining of AI models.
• Building AI models for the automatic segmentation of the prostate.
Over the past few months, Quibim has focused on adapting and integrating the Quibim Precision annotation environment into the ProCAncer-I platform. In the project, 5% of the cases uploaded to the platform are being annotated through the segmentation of prostate gland and the lesions. For this purpose, the functionalities of the annotation environment developed by Quibim are being used. This tool includes some manual annotation functionalities, such as the brush tool, which allows precise delineation of the regions of interest. In addition, for the segmentation of the prostate gland, three different areas are being delineated, including the central/transitional zone, the peripheral zone and the seminal vesicles. Segmenting all these structures from scratch is a time-consuming task. To speed up this segmentation, Quibim has integrated one of the functionalities of its QP-Prostate product, the algorithm for the automatic prostate segmentation. This algorithm generates a pre-segmentation that is reviewed by the radioligists of the consortium who perform its correction for final upload to ProstateNET All these annotations are stored in DICOM Seg, a standard format of the DICOM standard which allows storing in a single file a binary map of all the annotations made on a specific series, plus additional metadata such as the type of annotation (automatic, semi-automatic or manual), the author or the date together with the reference to the specific series, imaging study and patient. This facilitates the findability, interoperability and reusability (FAIR principles) of these annotations.